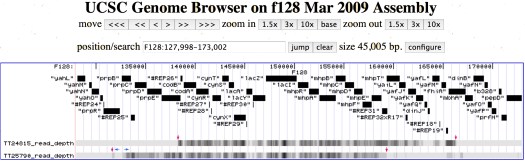
We can calculate levels of amplification as well as plasmid ploidy in a straightforward fashion from Solexa data. Consider our analysis of TT25790, which contains Elisabeth’s array EK568, fully characterized at both join points.
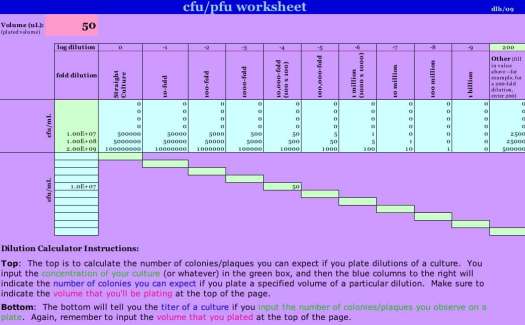
When we look at the “read density” map for reads crossing plasmid F’128(FC40) in this strain, we see that the frequency of reads increases abruptly at reference position ~132098 and returns to baseline abruptly at ~158297. If we go to the raw read data, we find 504973 reads in this interval of 26200 bp, for an average read density of 504973/26200 = 19.3 reads/nucleotide.
We can calculate in a similar manner the read density for the remaining, unamplified region of the plasmid, a circle of size 231427 bp. The total reads for the entire plasmid are 1569700, and for the unamplified region, 1569700-504973 = 1064727. Thus, the average unamplified read density will be 1064727/(231427-26200) = 5.2 reads/nucleotide.
Thus, the EK568 array is amplified with respect to the remainder of the plasmid by a factor of 19.3/5.2 = 3.7. This seems unusually low, given the fact that the samples were grown in minimal lactose medium. Even though the strain is rec, we expect this number to differ from preparation to preparation, as rec-independent recombination by mechanisms such as annealing, snap-back extension, and strand switching appears to be fairly frequent in F plasmid derivatives, perhaps the consequence of continuous rolling circle generation of long single-stranded DNA ends.
Returning to the raw data, we can ask what the read density across the 4068 bp lacIZ fusion gene itself is. We find 88003 reads across the gene, for a density of 88003/4068 = 21.6, which yields an amplification level of 21.6/5.2 = 4.2, still lower than expected.
Now, let’s look at the copy number of the plasmid itself. The chromosome contains 4857432 bp, across which we gathered 15716836 reads for an average density of 15716836/4857432 = 3.2 reads/nucleotide. We know that the unamplified region of the plasmid has a density of 5.2. Therefore, the ploidy of this plasmid with respect to the chromosome is 5.2/3.2 = 1.6, a trifle smaller than our working estimate of 2. Bear in mind, though, that this sample came from an overnight culture in stationary state. Under these conditions, we expect the copy number of F to be at its lowest.
We can use the read densities of the lacIZ gene and the chromosome to determine the copy number of the fusion per chromosome, equal to 21.5/3.2 = 6.8. If the activity of the mutant gene is 2% of wild-type and we assume strict additivity of gene expression, we calculate about 2×6.8 = 13.6 % final wild-type activity.
This may be enough to allow significant growth, but why wasn’t a greater growth rate selected by simple continued amplification? The answer may simply be that rec-independent amplification is slow compared to the relatively brief time to grow to stationary state with an already appreciable amount of lac activity.
Finally, why is the amplification level of lacIZ greater than the average amplification level of the array itself? The answer lies in the word “average”. Remember that this is a TID array in which the elements can be of different sizes. If many of the elements containing lacIZ are smaller than average, the actual level of the gene would be correspondingly greater, as observed.
Many thanks to Yong Lu for helping me collate this data!